Information Management and Preservation
Information and Software Engineering Group (IFS)Institute of Software Technology and Interactive Systems (ISIS)
Vienna University of Technology
Chemical Structure Recognition task 2012
Input | Output | Details | Training data | Test data | Tools | Contact
Last year, the Image2Structure task of TREC-CHEM invited participants to provide chemical structure files given a raster image of a chemical compound. The results were very positive, and this year we go one step further and make it more useful for the practitioners.
The 2012 CLEF-IP chemical image task gives you a full patent document as a PDF or TIF and asks you to 1. extract the locations of the chemical images in this document and 2. transform the so-recognized images into structure files.
Input
Tiff images containing full patent documents.
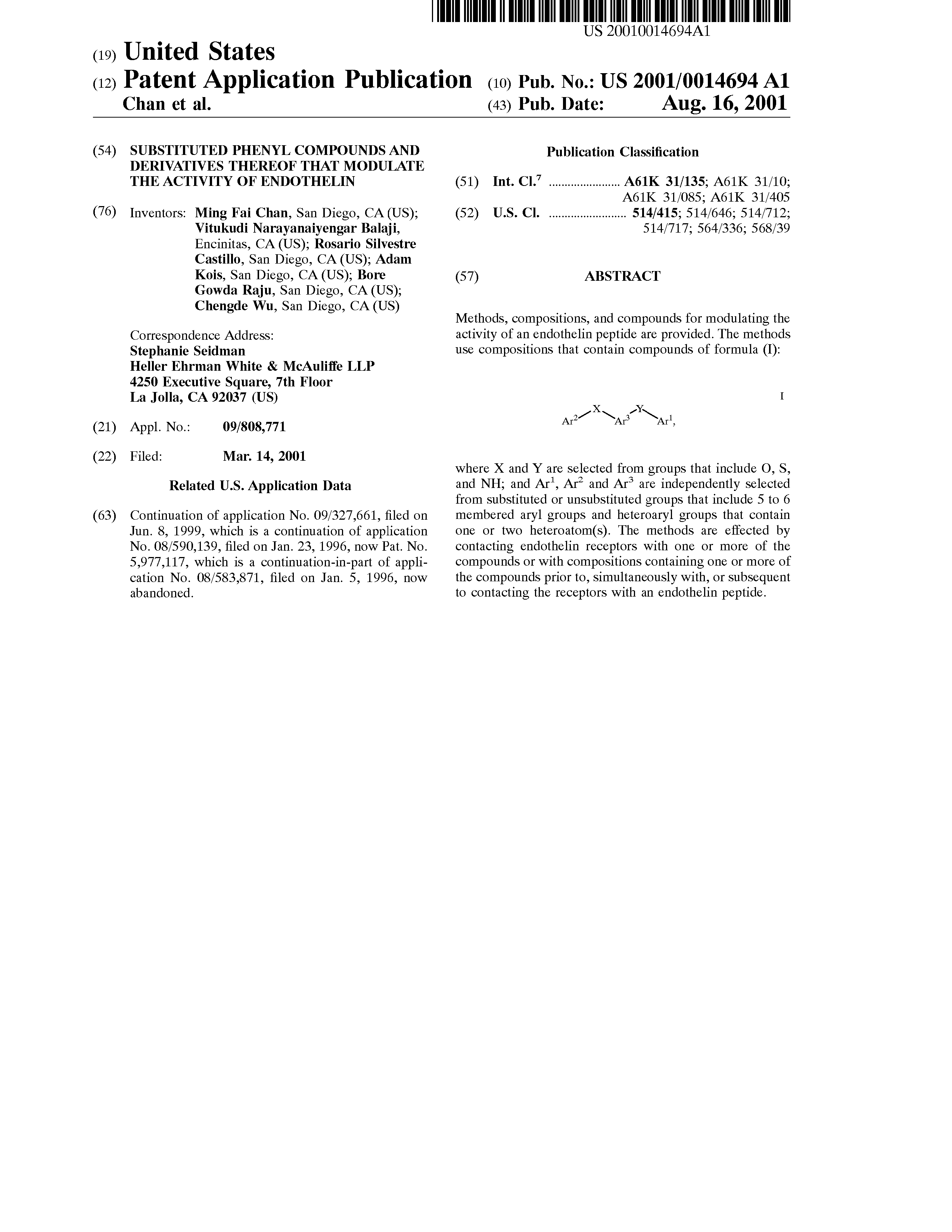
In order to make sure that those participants who may not have the resources to extract chemical images from the full-page images also participate, we will also make available a set of already extracted chemical images, similarly to the way the Image-to-Structure task had done in 2011.
Output
For each given patent document (e.g. US-20010014694-A1), you are to return a comma separated values (CSV) file listing a set of bounding boxes that identify the location of chemical images in the patent. Specifically, this file will be a sequence of lines, each line containing:- the name of the patent file
- the page number on which the current chemical image bounding box is located. Page numbering starts with 1
- the x-coordinate of the top left corner of the bounding box on the page. The coordinates of the page start at point (0,0) on the upper left corner and are counted in pixels. The coordinates are therefore such that the top left pixel of each page is (0,0), the x coordinate increases to the right (as usual) and the y coordinate increases downwards.
- the y-coordinate of the top left corner of the bounding box on the page.
- the width of the bounding box
- the height of the bounding box
Example of output file:
Contents of the CSV file:
US-20010014694-A1,1,1505,1085,295,52 US-20010014694-A1,4,1505,1151,295,52 US-20010014694-A1,5,290,2486,328,257 ...
For the set of already extracted images we will have provided as an input (technically, this should be a subset of what participants will have extracted in the first phase), the corresponding MOL files.
Example of Image-to-Structure recognition input and output
Chemical images
US20010014694A1_p0001_x1505_y1085_c00000.tif: US20010014694A1_p0004_x1505_y1151_c00001.tif:
US20010014694A1_p0004_x1505_y1151_c00001.tif:
 US20010014694A1_p0005_x0290_y2486_c00003.tif:
US20010014694A1_p0005_x0290_y2486_c00003.tif:
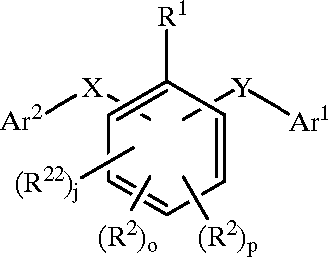
Chemical structures
US20010014694A1_p0001_x1505_y1085_c00000.molUS20010014694A1_p0004_x1505_y1151_c00001.mol
US20010014694A1_p0005_x0290_y2486_c00003.mol
Details
This tasks aims to bring the evaluation closer to practice and is therefore structured in two, rather disctinct, parts: chemical image extraction and chemical image recognition. Participants may submit results to only one of them. As can be seen below in the training data section, we will also provide a set of images already extracted from the patent files, for those participants who do not have the resources to do the extraction themselves, but can do the recognition part. The image below illustrates your task.
Evaluation
The evaluation of the two steps of this task will be done independently. Details about how exactly this will be done are still under discussion.Training data
Bounding box extraction
Here you have a set of 30 patents and manually extracted image clips.
Structure recognition
Here you have a subset of the images extracted above, with the corresponding MOL files. The images have been selected to be manageable, but still contain Markush structures - typical of patent chemical images.Test data
Bounding box extraction
Here you have a set of patent files for which you need to extract the bounding boxes of all chemicals.
Structure recognition
Here you have a set of tif files for which you need to provide MOL structure files, similarly to last year's image-to-structure recognition task at TREC-CHEM.
Tools
The segmentation results evaluation program is also available here. It includes a single .java file, a README.txt that explains everything (I hope) and a tiny sample ground truth and evaluation set file just to check that it has compiled and is working properly. Thanks to Alan Sexton for writing it.
The structure recognition task uses a perl scrip and the Chemistry::OpenBabel to identify the structure your method returns and compare it against the existing MOL file. However, this method works only for those structure for which InChi can be generated. For the training data, this means 94 out of the 133 pairs we provided. For the rest, the verification has to be done manually. Here is the script.
Contact
For questions and anything else regarding this task, please contact the organizers at (clef-ip-chem at ifs.tuwien.ac.at)

