Andreas Rauber
Institut für Softwaretechnik, Technische Universität Wien
Resselgasse 3/188, A-1040 Wien, Austria
http://www.ifs.tuwien.ac.at/ andi
Keywords: Data Visualization, Neural Networks, Dimensionality Reduction, Text Data Mining, Cluster Analysis
Real-world data is often represented in high-dimensional spaces and hence the similarities are hard to recognize. With the massive advance of huge data collections, tools for analyzing this data in order to extract and understand the hidden information become increasingly important. Particularly neural networks offer themselves for these data exploration tasks due to their capability of dealing with large volumes of noisy data. One of the most prominent neural networks for cluster analysis is the self organizing map (SOM) [4,5]. It provides a mapping from a high-dimensional feature space onto a usually 2-dimensional output space while preserving the topology of the input data as faithfully as possible. This characteristic is the reason why the SOM found large attraction in a wide range of application arenas.
However, in spite of all its benefits, its application has been limited by some drawbacks in terms of the interpretability of a trained SOM. Reading and interpreting the structure and the characteristics learned during the training process is not easily possible from the map display without expensive manual interaction. While the map is able to learn the distinctions between various clusters, their exact extent as well as their characteristics cannot be told from the standard SOM representation. This problem has led to the development of a number of enhanced visualization techniques supporting the interpretation of the self-organizing map. However, while these enhanced visualization techniques may provide assistance in identifying the cluster structure and cluster boundaries by manual inspection, they still do not give any information on the characteristics of the clusters. Thus, it still remains a tedious task to manually label the map, i.e. to determine the features that are characteristic for a particular cluster. Given an unknown data set that is mapped onto a self-organizing map, even with the visualization of clear cluster boundaries it remains a non-trivial task to elicit the features that are the most relevant and determining ones for a group of input data to form a cluster of its own, which features they share and which features distinguish them from other clusters. What we would like to have is a method which automatically labels a self-organizing map based on the features learned during the training process.
In this paper we present our novel LabelSOM approach to the automatic labeling of trained self-organizing maps based on the information provided by a trained SOM. In a nutshell, every unit of the map is labeled with the features that best characterize all the data points which are mapped onto that particular unit. This is achieved by using a combination of the quantization error of every feature and the relative importance of that feature in the weight vector of the unit. We demonstrate the benefits of this approach by labeling a SOM that was trained with a widely used reference data set describing animals by various attributes. The resulting labeled SOM gives a description of the animals mapped onto units and characterizes the various (sub)clusters present in the data set. We further provide a real-world example from the field of full text information mining based on a digital library SOM trained with the abstracts of scientific publications. This SOM thus represents a map of the scientific publications with the labels serving as a description of their topics and thus the various research fields.
The remainder of the paper is organized as follows: In Section II we present a brief review of related work on the interpretation, visualization and labeling of the self-organizing map. For the experiments reported in this paper we use two different data sets which are described in Section III. We then give a brief introduction to the self-organizing map, its architecture and training rule, presenting the results of applying the SOM to the analysis of the data sets in Section IV. Next, we introduce the LabelSOM method to automatically assign a set of labels for every unit in a trained SOM and provide results for both data sets in Section V. We further demonstrate how additional information on the cluster structure can be derived from the information provided by the labeling. A discussion of the presented LabelSOM method as well as its importance for the area of data mining is provided in Section VI. Finally, our conclusions are contained in Section VII.
Much progress has been made with respect to visualizing the cluster structure of a trained self-organizing map. Enhancing the standard representation, the U-Matrix [15] provides a kind of 3-dimensional visualization of the cluster distances between neighboring units, representing cluster boundaries as high ridges, or, in a corresponding 2-dimensional representation, as colored 2-d mappings of a 3-d landscape similar to conventional map representations.
A similar method allowing the interactive exploration of the distances between neighboring units in a trained SOM is provided by the Cluster Connection technique [8,9]. A set of thresholds can be set interactively in order to create a net of connected units allowing the exploration of intra and inter cluster similarities.
A different approach is being followed with the Adaptive Coordinate method [8,10,12], where the movement of the weight vectors of the SOM during the training process is mirrored in a 2-dimensional output space. Thus, clusters on the resulting map can be identified as clusters of units in the 2-dimensional AC representation of the map.
Although the combination of these methods provides a set of sophisticated tools for the analysis of the classification results of a trained SOM, no information on the characteristics of specific clusters can be deducted from the resulting representations. We may be able to identify clear cluster structures, yet have no way to tell, which characteristics makes the clusters stand apart. The map itself simply represents a two-dimensional plane where data points are located according to their overall similarity, with no information on their specific similarities and dissimilarities available. Thus, for intuitive representation, a trained SOM needs to be labeled.
So far the majority of SOMs is labeled manually, i.e. after inspecting a trained map, descriptive labels are assigned to specific regions in a map. While this is a perfectly suitable approach for the labeling of small maps where some knowledge about the underlying data is present, it is unfeasible with large maps of high dimensionality and unknown data characteristics.
Quite frequently we also find a SOM to be labeled directly with the labels of the input data mapped onto each particular unit. This provides a good overview of the resulting mapping as long as the labels of the data points convey some information on their characteristics. In many situations, especially in the area of data mining, this presumption does not hold, with the labels of the data points often simply being enumerations of the data sample.
In some situations preclassified information is available such as in the Websom project [2], where the units of a SOM representing Usenet newsgroup articles are labeled with the name of the newsgroup or newsgroup hierarchy that the majority of articles on a unit comes from. This allows for a kind of automatic assignment of labels to the units of a SOM using the additional knowledge provided by the preclassification of articles in newsgroups.
A method using component planes for visualizing the contribution of each variable in the organization of a map has been presented recently [3]. The individual weight vector elements of the map are considered as separate component planes which can be visualized independently similar to the U-Matrix method for SOM representation. By manual inspection this provides some additional information on coherent regions for each vector component. However, it requires manual interaction by examining each dimension separately and does thus not offer itself to automatic labeling of SOMs trained with high-dimensional input data.
What we would like to have is a way to automatically label the units of a SOM based on the characteristics learned during the training process.
For the experiments presented hereafter we use 2 datasets. First, we present the principles of the LabelSOM method using the Animals data set [13], a well-known reference example, which has frequently been used to demonstrate the clustering capabilities of the self-organizing map, c.f. [1,10]. In this data set 16 animals are described by 13 attributes as given in Table I. Please note that, contrary to the experiments described in [13], we did not encode the animals names, resulting in two pairs of vectors being identical. In this dataset we can basically identify 2 clusters of animals, namely birds and mammals, strongly separated by their number of legs as well as the fact whether they have feathers or fur. Within these two clusters further subclusters can be identified based on their size, looks and habits.
|
Following this reference example, we present the benefits of the LabelSOM method using a real-world example from the field of text mining based on scientific abstracts.
For this we use a set of 48 abstracts from publications of our department.
These abstracts were transformed into a vector representation following the vector space model of information retrieval.
We used full-term indexing to extract a list of terms while applying some basic stemming rules.
Furthermore, we eliminated words that appear in more than 90% or less than 3 abstracts, with this rule saving us from having to specify language or content specific stop word lists.
Using an absolute number of 3 abstracts for the minimum number of abstracts a word must be present in instead of specifying a percentage of the total number of abstracts allows us to set a desired granularity level of detail representation in the document vectors independent from the size of the dataset.
The documents are represented using a
![]() ,
i.e. term frequency
,
i.e. term frequency ![]() inverse document frequency weighting scheme [14].
This weighting scheme assigns high values to terms that are considered important in terms of describing the contents of a document and discriminating between the various abstracts.
For the 48 abstracts the indexing process identified 482 content terms which are used for SOM network training to produce a clustering of the abstracts in terms of their contents.
inverse document frequency weighting scheme [14].
This weighting scheme assigns high values to terms that are considered important in terms of describing the contents of a document and discriminating between the various abstracts.
For the 48 abstracts the indexing process identified 482 content terms which are used for SOM network training to produce a clustering of the abstracts in terms of their contents.
The self-organizing map is an unsupervised neural network providing a mapping from a high-dimensional input space to a usually two-dimensional output space while preserving topological relations as faithfully as possible.
The SOM consists of a set of i units arranged in a two-dimensional grid, with a weight vector
![]() attached to each unit.
Elements from the high dimensional input space, referred to as input vectors
attached to each unit.
Elements from the high dimensional input space, referred to as input vectors
![]() ,
are presented to the SOM and the activation of each unit for the presented input vector is calculated using an activation function.
Commonly, the Euclidean distance between the weight vector of the unit and the input vector serves as the activation function.
In the next step the weight vector of the unit showing the highest activation (i.e. the smallest Euclidean distance) is selected as the `winner' and is modified as to more closely resemble the presented input vector.
Pragmatically speaking, the weight vector of the winner is moved towards the presented input signal by a certain fraction of the Euclidean distance as indicated by a time-decreasing learning rate
,
are presented to the SOM and the activation of each unit for the presented input vector is calculated using an activation function.
Commonly, the Euclidean distance between the weight vector of the unit and the input vector serves as the activation function.
In the next step the weight vector of the unit showing the highest activation (i.e. the smallest Euclidean distance) is selected as the `winner' and is modified as to more closely resemble the presented input vector.
Pragmatically speaking, the weight vector of the winner is moved towards the presented input signal by a certain fraction of the Euclidean distance as indicated by a time-decreasing learning rate ![]() .
Thus, this unit's activation will be even higher the next time the same input signal is presented.
Furthermore, the weight vectors of units in the neighborhood of the winner as described by a time-decreasing neighborhood function
.
Thus, this unit's activation will be even higher the next time the same input signal is presented.
Furthermore, the weight vectors of units in the neighborhood of the winner as described by a time-decreasing neighborhood function ![]() are modified accordingly, yet to a less strong amount as compared to the winner.
This learning procedure finally leads to a topologically ordered mapping of the presented input signals.
Similar input data is mapped onto neighboring regions on the map [5].
are modified accordingly, yet to a less strong amount as compared to the winner.
This learning procedure finally leads to a topologically ordered mapping of the presented input signals.
Similar input data is mapped onto neighboring regions on the map [5].
Figure 1 depicts the result of training a
![]() SOM with the animal-data provided in Table I.
Following the training procedure we find, that animals exhibiting similar characteristics are mapped close to each other on the resulting SOM.
In the standard representation depicted in Figure 1, the units are represented by the rectangles in the map, with each unit being assigned the names of those input vectors that were mapped onto it, i.e. for which it was the winner.
Judging from the names of the feature vectors, we find that the upper half of the map represents solely birds, while all mammals are mapped onto the lower half of the map.
We furthermore find a distinction between hunting animals in the left area of the map as opposed to non-hunting animals to the right.
Further clusters may be identified using any of the enhanced cluster visualization techniques presented before.
However, we are only able to do this type of interpretation for the resulting SOM representation because of our knowledge on the underlying data and because of its low dimensionality of 13 attributes.
SOM with the animal-data provided in Table I.
Following the training procedure we find, that animals exhibiting similar characteristics are mapped close to each other on the resulting SOM.
In the standard representation depicted in Figure 1, the units are represented by the rectangles in the map, with each unit being assigned the names of those input vectors that were mapped onto it, i.e. for which it was the winner.
Judging from the names of the feature vectors, we find that the upper half of the map represents solely birds, while all mammals are mapped onto the lower half of the map.
We furthermore find a distinction between hunting animals in the left area of the map as opposed to non-hunting animals to the right.
Further clusters may be identified using any of the enhanced cluster visualization techniques presented before.
However, we are only able to do this type of interpretation for the resulting SOM representation because of our knowledge on the underlying data and because of its low dimensionality of 13 attributes.
Figure 2, for example, represents a 7 ![]() 7 SOM trained with the scientific abstracts data.
In this application, the SOM is intended to provide a clustering of the documents based on contents similar to the organization of documents in a conventional library.
(For some deeper discussion on the utilization of SOMs in text data mining we refer to [6,7,11].)
Again, the units are labeled with the names of the document vectors, which consist of the first 3 letters of the author's name followed by the short name of the conference or workshop the paper was published at.
Without any additional knowledge on either the conferences or the authors, the given representation is hard to interpret although we might draw some conclusions on the cluster structure by considering the authors names as indicators.
Enhanced visualization techniques again would help us in detecting the cluster structure while albeit providing us with no information on the content of the map, i.e. the characteristics of the clusters.
The only way to interpret this SOM requires us to read all the abstracts in order to identify descriptive terms for the various units and regions.
7 SOM trained with the scientific abstracts data.
In this application, the SOM is intended to provide a clustering of the documents based on contents similar to the organization of documents in a conventional library.
(For some deeper discussion on the utilization of SOMs in text data mining we refer to [6,7,11].)
Again, the units are labeled with the names of the document vectors, which consist of the first 3 letters of the author's name followed by the short name of the conference or workshop the paper was published at.
Without any additional knowledge on either the conferences or the authors, the given representation is hard to interpret although we might draw some conclusions on the cluster structure by considering the authors names as indicators.
Enhanced visualization techniques again would help us in detecting the cluster structure while albeit providing us with no information on the content of the map, i.e. the characteristics of the clusters.
The only way to interpret this SOM requires us to read all the abstracts in order to identify descriptive terms for the various units and regions.
With no a priori knowledge on the data, even providing information on the cluster boundaries does not reveal information on the relevance of single attributes for the clustering and classification process. In the LabelSOM approach we determine those vector elements (i.e. features of the input space) that are most relevant for the mapping of an input vector onto a specific unit. This is basically done by determining the contribution of every element in the vector towards the overall Euclidean distance between an input vector and the winners' weight vector, which forms the basis of the SOM training process.
The LabelSOM method is built upon the observation, that, after SOM training, the weight vector elements resemble as far as possible the corresponding input vector elements of all input signals that are mapped onto this particular unit as well as to some extent those of the input signals mapped onto neighboring units.
Vector elements having about the same value within the set of input vectors mapped onto a certain unit describe the unit in so far as they denominate a common feature of all data signals of this unit.
If a majority of input signals mapped onto a particular unit exhibit a highly similar input vector value for a particular feature, the corresponding weight vector value will be highly similar as well.
We can thus select those weight vector elements, which show by and large the same vector element value for all input signals mapped onto a particular unit to serve as a descriptor for that very unit.
The quantization error for all individual features serves as a guide for their relevance as a class label.
We select those vector elements that exhibit a quantization error vector element value of close to 0.
The quantization error vector is computed for every unit i as the accumulated distance between the weight vector elements of all input signals mapped onto unit i and the unit's weight vector elements.
More formally, this is done as follows:
Let Ci be the set of input patterns
![]() mapped onto unit i.
Summing up the distances for each vector element k over all the vectors xj (
mapped onto unit i.
Summing up the distances for each vector element k over all the vectors xj (
![]() )
yields a quantization error vector qi for every unit i (Equation 1).
)
yields a quantization error vector qi for every unit i (Equation 1).
Selecting those weight vector elements that exhibit a corresponding quantization error of close to 0 thus results in a list of attributes that are shared by and large by all input signals on the respective unit and thus describe the characteristics of the data on that unit. These attributes thus serve as labels for regions of the map for data mining applications.
In text mining applications we are usually faced with a further restriction.
Due to the high dimensionality of the vector space and the characteristics of the
![]() representation of the document feature vectors, we usually find a high number of input vector elements that have a value of 0, i.e. there is a large number of terms that is not present in a group of documents.
These terms obviously yield a quantization error value of 0 and would thus be chosen as labels for the units.
Doing that would result in labeling the units with attributes that are not present in the data on the respective unit.
While this may be perfectly ok for some data analysis tasks, where even the absence of an attribute is a distinctive characteristics, it is definitely not the goal in text mining applications where we want to describe the present features that are responsible for a certain clustering rather than describe a cluster via the features that are not present in its data.
Hence, we need to determine those vector elements from each weight vector which, on the one hand, exhibit about the same value for all input signals mapped onto that specific unit as well as, on the other hand, have a high overall weight vector value indicating its importance.
To achieve this we define a threshold
representation of the document feature vectors, we usually find a high number of input vector elements that have a value of 0, i.e. there is a large number of terms that is not present in a group of documents.
These terms obviously yield a quantization error value of 0 and would thus be chosen as labels for the units.
Doing that would result in labeling the units with attributes that are not present in the data on the respective unit.
While this may be perfectly ok for some data analysis tasks, where even the absence of an attribute is a distinctive characteristics, it is definitely not the goal in text mining applications where we want to describe the present features that are responsible for a certain clustering rather than describe a cluster via the features that are not present in its data.
Hence, we need to determine those vector elements from each weight vector which, on the one hand, exhibit about the same value for all input signals mapped onto that specific unit as well as, on the other hand, have a high overall weight vector value indicating its importance.
To achieve this we define a threshold ![]() in order to select only those attributes that, apart from having a very low quantization error, exhibit a corresponding weight vector value above
in order to select only those attributes that, apart from having a very low quantization error, exhibit a corresponding weight vector value above ![]() .
In these terms,
.
In these terms, ![]() can be thought of indicating the minimum importance of an attribute with respect to the
can be thought of indicating the minimum importance of an attribute with respect to the
![]() representation to be selected as a label.
representation to be selected as a label.
Figure 3 shows the result of labeling the
![]() SOM trained with the animals data set depicted in Figure 1.
Each unit is assigned a set of up to 5 labels based on the quantization error vector and the unit's weight vector (
SOM trained with the animals data set depicted in Figure 1.
Each unit is assigned a set of up to 5 labels based on the quantization error vector and the unit's weight vector (
![]() ).
We find that each animal is labeled with its characteristic attributes, i.e. all birds are identified as having feathers and 2 legs whereas the mammals have 4 legs and hair.
The remaining subclusters are identified by the size of the animals and their preferences for hunting, flying, swimming, etc.
For example, the big mammals are located in the lower right corner of the map as a subcluster of the mammals.
As another subcluster consider the distinction of hunting vs. non-hunting animals - irrespective of their belonging to the group of birds or group of mammals.
The hunting animals by and large may be found on the left side of the map whereas the non-hunting animals are located on the right side.
Thus, we can not only identify the decisive attributes for the assignment of every input signal to a specific unit but also detect the cluster boundaries and tell the characteristics and extents of subclusters within the map.
Mind, that not all units have the full set of 5 labels assigned, i.e. one or more labels are empty (none) like e.g. for the unit representing the dog in the lower left corner.
This is due to the fact that less than 5 vector elements have a weight vector value mik greater than
).
We find that each animal is labeled with its characteristic attributes, i.e. all birds are identified as having feathers and 2 legs whereas the mammals have 4 legs and hair.
The remaining subclusters are identified by the size of the animals and their preferences for hunting, flying, swimming, etc.
For example, the big mammals are located in the lower right corner of the map as a subcluster of the mammals.
As another subcluster consider the distinction of hunting vs. non-hunting animals - irrespective of their belonging to the group of birds or group of mammals.
The hunting animals by and large may be found on the left side of the map whereas the non-hunting animals are located on the right side.
Thus, we can not only identify the decisive attributes for the assignment of every input signal to a specific unit but also detect the cluster boundaries and tell the characteristics and extents of subclusters within the map.
Mind, that not all units have the full set of 5 labels assigned, i.e. one or more labels are empty (none) like e.g. for the unit representing the dog in the lower left corner.
This is due to the fact that less than 5 vector elements have a weight vector value mik greater than ![]() .
Another interesting fact can be observed on unit (5/2)
.
Another interesting fact can be observed on unit (5/2)![]() , which is not winner for any input signal and is labeled with small, 2 legs, big, 4 legs, hair, obviously representing a mixture of mammals and birds and thus exhibiting both characteristics.
, which is not winner for any input signal and is labeled with small, 2 legs, big, 4 legs, hair, obviously representing a mixture of mammals and birds and thus exhibiting both characteristics.
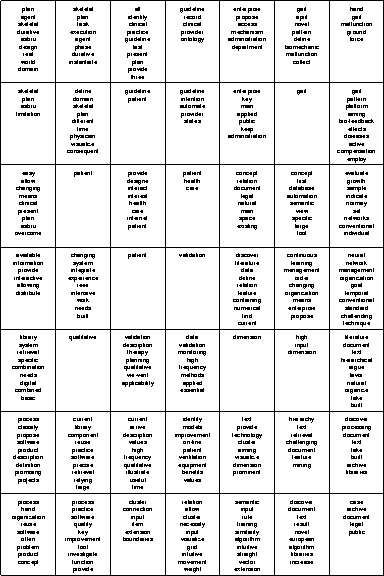
Figure 4 depicts the
![]() SOM given in Figure 2, with this time having a set of up to 10 labels automatically assigned to the the units.
It leaves us with a clearer picture of the underlying text archive and allows us to understand the reasons for a certain cluster assignment as well as identify topics and areas of interest within the document collection.
For example, in the upper left corner we find a group of units sharing labels like skeletal plans, clinical, guideline, patient, health which deal with the development and representation of skeletal plans for medical applications.
Another homogeneous cluster can be found in the upper right corner which is identified by labels like gait, pattern, malfunction and deals with the analysis of human gait patterns to identify malfunctions.
A set of units in the lower left corner of the map is identified by a group of labels containing, among others, software, process, reuse and identifies papers dealing with software reuse.
This is followed by a large cluster to the right labeled with cluster, intuitive, document, archive, text, input containing papers on the problems of cluster visualization and its application in the context of document archives.
Further clusters can be identified in the center of the map on plan validation, and quality analysis, neural networks etc.
The map is available for further interactive exploration at http://www.ifs.tuwien.ac.at/ifs/research/ir/IFS_Abstracts/.
SOM given in Figure 2, with this time having a set of up to 10 labels automatically assigned to the the units.
It leaves us with a clearer picture of the underlying text archive and allows us to understand the reasons for a certain cluster assignment as well as identify topics and areas of interest within the document collection.
For example, in the upper left corner we find a group of units sharing labels like skeletal plans, clinical, guideline, patient, health which deal with the development and representation of skeletal plans for medical applications.
Another homogeneous cluster can be found in the upper right corner which is identified by labels like gait, pattern, malfunction and deals with the analysis of human gait patterns to identify malfunctions.
A set of units in the lower left corner of the map is identified by a group of labels containing, among others, software, process, reuse and identifies papers dealing with software reuse.
This is followed by a large cluster to the right labeled with cluster, intuitive, document, archive, text, input containing papers on the problems of cluster visualization and its application in the context of document archives.
Further clusters can be identified in the center of the map on plan validation, and quality analysis, neural networks etc.
The map is available for further interactive exploration at http://www.ifs.tuwien.ac.at/ifs/research/ir/IFS_Abstracts/.
 |
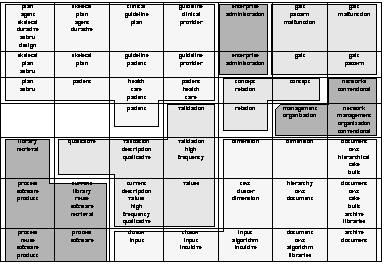 |
Additionally to the increased interpretability, cluster detection is facilitated using the labels derived from the LabelSOM method.
Clear cluster boundaries can be defined by combining units sharing a set of labels.
These shared labels can then be used as higher-level class identifiers.
Applied to the 7 ![]() 7 SOM representing the scientific abstracts, this results in a total of 8 different clusters with a smaller number of cluster labels as shown in Figure 5.
7 SOM representing the scientific abstracts, this results in a total of 8 different clusters with a smaller number of cluster labels as shown in Figure 5.
While the labels identified by our LabelSOM method in the text data mining example can probably not serve directly as class labels in the conventional sense, they reveal a wealth of information about the underlying map and the structures learned during the self-organizing training process. The user gets a justification for the clustering as well as information on the sub-structure within clusters by the very attributes.
The labels themselves aid in identifying the most important features within every unit and thus help to understand the information represented by a particular unit. In spite of the little redundancy present in abstracts, the labels turn out to be informative in so far as they help the user to understand the map and the data set as such. Especially in cases where little to no knowledge on the data set itself is available, the resulting representation can lead to tremendous benefits in understanding the characteristics of the set as a whole as well as of individual data items. Apart from data mining purposes they can serve as a basis for simplified semi-automatic creation of class labels by allowing the user to choose the most appropriate terms from the automatically created list.
It is important to mention that the information used for the labeling originates entirely from the self-organizing process of the SOM without the use of sophisticated machine learning techniques. With the increasing use of self-organizing maps in the data mining area, the automatic labeling of maps to identify the features of certain clusters based on the training process itself becomes an important aid in correctly applying the process and interpreting the results. Being based on a neural network approach with high noise tolerance allows the application of the LabelSOM approach in a wide range of domains, especially in the analysis of very high-dimensional input spaces.
We have presented the LabelSOM method to automatically assign labels to the units of a trained self-organizing map. This is achieved by determining those features from the high-dimensional feature space that are most relevant for a certain input data to cluster assignment. The quantization error for every vector element is calculated by taking the weighted sum of the activation function for all input signals mapped onto a specific unit and selecting those vector elements with the highest internal similarity as unit labels. The resulting benefits are twofold: First, assigning labels to each unit helps with the interpretation of single clusters by making the common features of a set of data signals that are mapped onto the same unit explicit. This serves as a description for each set of data mapped onto a unit. Second, by taking a look at groups of neighboring units sharing common labels it is possible to determine sets of units forming larger clusters, to identify cluster and sub-cluster boundaries and to provide specific information on the differences between certain clusters. Last, but not least, labeling the map allows it to be actually read.
This document was generated using the LaTeX2HTML translator Version 98.1p1 release (March 2nd, 1998)
Copyright © 1993, 1994, 1995, 1996, 1997, Nikos Drakos, Computer Based Learning Unit, University of Leeds.
The command line arguments were:
latex2html -split 0 ijcnn99.tex.
The translation was initiated by Andreas RAUBER on 1999-08-19